Dr Robb Hollis, University of Edinburgh
Ovarian cancer is a leading cause of female cancer deaths.1 High grade serous ovarian carcinoma (HGSOC) is by far the most common form of the disease (70% of cases).2 While HGSOC is usually highlysensitive to initial platinum-based therapy, recurrence is common and accumulates therapy resistance (chemoresistance), leading to treatment failure.2 Acquired chemoresistance therefore represents the single greatest challenge in ovarian cancer management. Identifying molecular drivers of chemoresistance affords the opportunity to uncover new therapeutic targets that could prevent or reverse chemoresistance acquisition.
MicroRNAs are small, single-stranded non-coding RNAs that function to post-transcriptionally regulate gene expression.3 Recent findings from our group have identified microRNAs as potential mediators of epithelial-to-mesenchymal transition (EMT) in ovarian cancer,4 which has been associated with acquired chemoresistance in ovarian cancers and other tumour types.5-7 These findings were based on a novel approach of using ovarian carcinosarcoma (OCS) (Figure 1) – a rare and intrinsically chemoresistant form of ovarian cancer that harbours both carcinomatous and sarcomatous components8 – as a model of EMT and chemoresistance.4
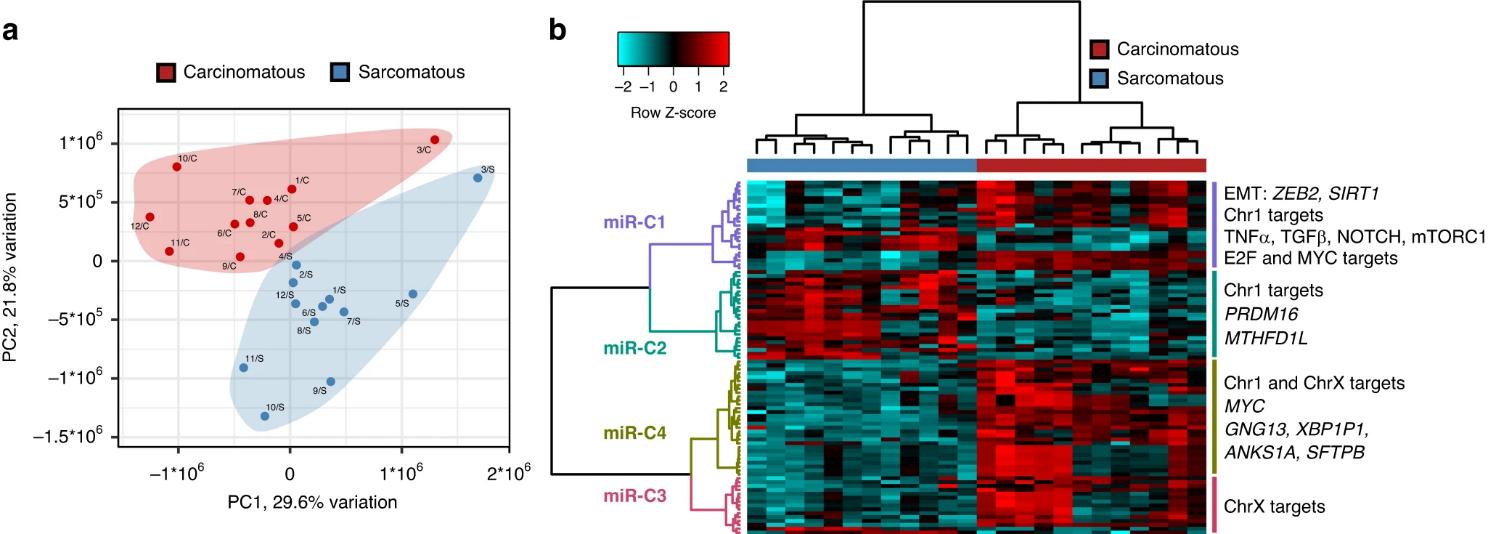
Figure 1. Analysis of the two tumour compartments in ovarian carcinosarcoma reveals global changes in microRNA profiles. (a) Global microRNA expression patterns separates tumour compartments from multiple patients, (b) many microRNAs are highlydifferentially expressed between carcinomatous and sarcomatous compartments. From Herrington et al. Brit J Cancer 20244
This project aims to investigate whether these candidate chemoresistance-driving microRNAs:
(i) Are associated with chemoresistance acquisition in HGSOC
(ii) Can drive acquisition of chemoresistance in laboratory models of HGSOC
(iii) Activate downstream therapeutically-targetable chemoresistance-driving pathways
The PhD candidate working on this project will develop a mixed wet laboratory and computation (bioinformatic) skillset using the following resources available at the CRUK Scotland Centre:
• An extensive cohort of matched diagnosis-relapse (temporally-separated) HGSOC samples
• A cohort of samples from multiple sites at HGSOC diagnosis (spatially-separated samples)
• Access to new cohorts of retrospective HGSOC cases with samples from diagnosis
• The largest OCS tissue resource available anywhere in the world
• A library of ovarian cancer cell models representing the breadth of molecular subtypes
• A collection of primary patient-derived ovarian cancer models established locally at the centre
This project will enable the successful candidate to develop a broad portfolio of cancer research techniques, including experience in both tissue-based and in vitro cancer studies. The individual will be supported by a vibrant research team at The Nicola Murray Centre for Ovarian Cancer Research at The University of Edinburgh, comprising a large number of postdoctoral scientists, clinician scientists, technicalstaff and PhD students. They will also receive additional support from an extensive network of researchers across Scotland via the Cancer Research UK Scotland Centre.
For further information on the project or informal enquiries, please contact Dr Robb Hollis, This email address is being protected from spambots. You need JavaScript enabled to view it..
To place an application, please visit this site at the University of Edinburgh.
When submitting your application please make sure that you have also completed your application to the Windsor Fellowship and please upload the completed recruitment form.
Duration: 4 years, starting October 2026
Closing Date: 24th November 2025
Interview for this position will take place in January 2026
Lab Websites
Dr Robb Hollis - Molecular defects in Ovarian Cancer
References
1. Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263.
2. Hollis RL. Molecular characteristics and clinical behaviour of epithelial ovarian cancers. Cancer Lett. 2023;555:216057.
3. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18.
4. Herrington CS, Oswald AJ, Stillie LJ, Croy I, Churchman M, Hollis RL. Compartment-specific multiomic profiling identifies SRC and GNAS as candidate drivers of epithelial-to-mesenchymal transition in ovarian carcinosarcoma. Br J Cancer. 2024;130:327-335.
5. Fischer KR, Durrans A, Lee S, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472-476.
6. Zheng X, Carstens JL, Kim J, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525-530.
7. Loret N, Denys H, Tummers P, Berx G. The Role of Epithelial-to-Mesenchymal Plasticity in Ovarian Cancer Progression and Therapy Resistance. Cancers. 2019;11.
8. Hollis RL, Croy I, Churchman M, et al. Ovarian carcinosarcoma is a distinct form of ovarian cancer with poorer survival compared to tubo-ovarian high-grade serous carcinoma. Br J Cancer. 2022;127:1034-1042.