Enabling infrastructure is central to the success of our Centre. We have identified 4 priority areas for enhanced funding to unblock restrictions in the translational pipeline to ensure our discovery science leads to patient benefit: Tissue Resources and Digital and Molecular Pathology; Preclinical Models of Cancer; Advanced Imaging for Research; and Data Science: Data Gathering, Provision and Analysis.
Tissue Resources and Digital & Molecular Pathology
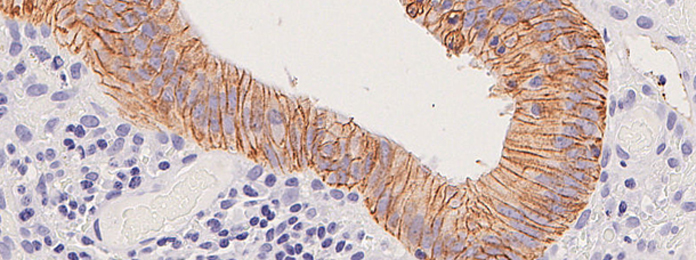
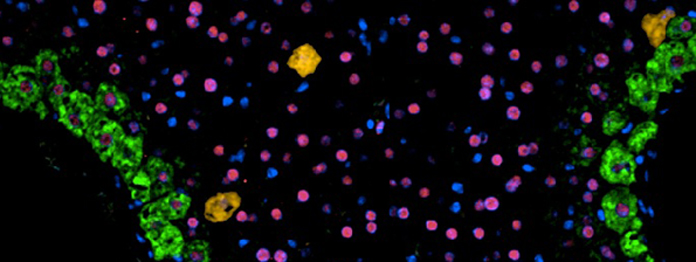
Tissue Resources and Digital and Molecular Pathology enable tissue collection; archival sample identification; morphological assessment; image capture, curation and storage; comparative pathology (largely mouse-human); automated immunohistochemistry, immunofluorescence and RNA in situ hybridisation, including multiplex analyses; spatial transcriptomics support; and digital pathology using QPath, Halo, and Visiopharm platforms. All samples, equipment and data will be shared across the Centre.
The Glasgow Tissue Research Facility (GTRF) works closely with the NHS Greater Glasgow & Clyde (NHSGGC) Biorepository, which, with appropriate governance in place, facilitates tissue collection and provides researchers with access to a wide variety of tissue including pathology archive samples, surplus diagnostic or surgical tissue and bespoke tissue collected from NHS Scotland patients as part of cohort studies or clinical trials. The GTRF provides bespoke tissue microarrays, routine H&E staining, digital pathology scanning and image analysis software to enable data integration. The Edinburgh facility works closely with NHS Lothian and is integrated into the Centre. Its activities are very closely mirrored, and all Centre specific SOPs will be harmonised across localities.
Crucially, both NHSGGC Biorepository and NHS Lothian Bioresource have the capability to perform focused sample collections of multiple tissues and fluids, including fresh and frozen samples and specialist media. They frequently liaise with other Scottish Health Boards for multi-centre collection, which we plan to fund where this is essential (e.g. population-based programmes, rare cancer types). We will endeavour to enhance our collection and interrogation of clinical samples from patients with detailed clinical real world data including tumour characteristics, therapy response, and recurrence and metastasis. Working closely with Experimental Cancer Medicine Centres (ECMCs), we maximise sample and data collection from our clinical trial patients.
One of our key aims is to generate detailed spatial maps of human cancer tissue, and to overlap these with our complex mouse models. This is essential to ensure that the key biological processes we study in our preclinical models reflect disease in patients and that we accurately understand how these processes are modulated by therapy. Furthermore, use of state-of-the-art imaging in our mouse models and in human tissue will allow us to make critical new insights into the biology and potential vulnerabilities of the metastatic niche and tumour microenvironment.
Molecular Pathology is increasingly available as a routine across Glasgow and Edinburgh cancer patients through programmes such as Precision-Panc and IMAGINE, and the Glasgow Precision Oncology Laboratory (GPOL), which routinely provides accredited DNA sequencing of a large driver gene panel for clinical use. As a first step to develop Scotland’s clinical genomic diagnostic capability, we aim to integrate genomic testing into routine care. GPOL will also provide not-for-profit panel sequencing for research and analyse outputs using their bespoke pipeline. Other nucleic acids research will be outsourced to accredited providers of Illumina, Nanpore and TempoSeq sequencing, and analysed by Centre bioinformatics groups.
Preclinical Models of Cancer
Working with the ECMCs, tissue banks, tissue research facilities and our wider collaborative network, we have developed pipelines and processes to build banks of human patient derived cell lines, patient derived organoids and patient derived xenografts. These include those from all of our focused tumour types representing different disease stages and subtypes and those derived pre and post therapeutic intervention. We will further invest to strengthen these pipelines.
One of the greatest challenges in cancer is to predict patient therapeutic outcomes. Towards this goal, a particular strength of the Centre will be our collection of preclinical models, especially complex, genetically engineered mouse (GEM) models. We have used these models over the last 5 years to support development of UK-wide networks to tackle hard-to-treat cancers: pancreatic cancer (Precision-Panc), hepatocellular cancer (HUNTER), mesothelioma (PREDICT-Meso), colorectal cancer (ACRCelerate) and glioma (GCGR: glioma cellular genetics resource). These models allow us to: understand the fundamental biology of cancer; effectively disease-position these models in terms of human cancer subtypes based on genetics, gene expression, pathology and outcomes to better mirror human disease; and identify and predict novel targets/combinations for therapeutic intervention. They also enable us to interrogate key markers of tumour development in the context of an immunocompetent organism, providing a rich resource for biological discovery and the development of therapeutic preclinical trial platforms. Our models include those of pre-malignant and early-stage disease, which will be useful for early detection, and of metastasis, which will help identify and target pre-metastatic and metastatic niches. For each of these cancers, we have set up strong translational/clinical partnerships within Glasgow and Edinburgh and more broadly Scotland. The utilisation and further development of these complex preclinical models of cancer will be a central component of the Centre.
In addition to GEM models, we have invested in expanding our capabilities to model complex human cancer genetics in the fly. Ross Cagan is utilising Drosophila to investigate cancer therapeutics and metastasis, particularly in colorectal cancer and has a great strength in drug discovery and chemical biology. Indeed, he has used Drosophila genetic approaches to identify drugs and new drug combinations that he has shown to be effective in clinical trials (Bangi et al. iScience 2021, Sci Adv 2019).
Liz Patton’s work in zebrafish models of melanoma has demonstrated genes that are critical for melanocyte stem cell regeneration (Johansson et al. Dev Cell 2020), has identified melanoma transcriptional subtypes (Travnickova et al. Cancer Res 2019) and has explored convergence of signalling pathways (Ngeow et al. PNAS 2018) and impact of mosaic RAS/MAPK variants (Al-Olabi et al. J Clin Invest 2018). We are expanding the use of zebrafish models of cancer beyond established melanoma models to those including brain and gynaecological cancer.
Advanced Imaging for Research
Edinburgh research imaging is provided by the institute within which each researcher is based. For example, the Advanced Imaging Facility in the IGC is partly Centre-funded and exceptionally well equipped with all routinely used modalities and more specialised platforms such as STORM, Optical Projection Tomography, high content imaging including Quantitative Pathology and super-resolution systems. We will also benefit considerably from new and enhanced imaging facilities provided by the HGU 4D cellular medicine centre within the IGC.
The Glasgow imaging facility is within the Scotland Institute. It has a wide range of specialised microscopes that provide users with the flexibility to approach their research using a diverse range of applications. Microscopes include a variety of confocal and wide-field systems, super-resolution systems, high-content screening, in vivo imaging systems, and equipment for advanced techniques such as TIRF, FLIM/FRET and multiphoton in vivo imaging. The Translational Molecular Imaging (TMI) facility advances novel imaging technologies and serves as a central hub for emerging whole animal and human imaging research and technology. The state-of-the-art facilities and equipment include access to a GE cyclotron, dedicated research radiosythesizers, a small animal PET/MRI facility and two clinical GE Discovery Time-of-flight PET/CT scanners. The focus is on developing new PET radiotracers and MRI methodology critical to understanding tumour metabolism and biology in vivo in mice and humans and serves as key infrastructure for the Centre’s ambitions.
Data Sciences: Data Gathering, Provision and Analysis
Data science supports the specific data-dependent activities of tumour research themes, and thereby acquire essential experience in obtaining research-useful data from Scottish oncology and cancer screening patients. To prevent over-reach in our population-based data activities and ambitions, colorectal cancer (CRC) and hepatobiliary cancer (HBP) will act as pathfinders. Anonymised patient data will be required for identifying high-risk patients, for optimising screening and surveillance, and for identifying patients for research and experimental medicine studies. The experience we thereby acquire will enable us efficiently to obtain high quality data for the other themes, for local research into other cancers, and for other cancer researchers in Scotland. We will throughout make use of the considerable local infrastructure and expertise in data storage, safe havens and AI-based analysis, and maintain contacts with other UK groups and CRUK-funded data initiatives.
It is critical that we work within and navigate the existing local data infrastructure. This is complex and we have mapped it as a preliminary project. Our strategy is to place Centre data wranglers into PHS, NHS Lothian DataLoch and NHS GGC Safe Haven to work with the teams there. We will work with the relevant data stores and safe havens to bring together Scotland-wide data (e.g. CRC screening) or Lothian-GGC data (e.g. patients for HBP surveillance) into a single Centre data haven hosted by EPCC, in which we can perform joint data analyses. These data will be updated periodically (for new patients and new episodes) and can also be shared more widely (e.g. with CRUK wide initiatives) subject to permissions. We shall also bring other available data into each theme haven, such as cancer genomics and research-only data. Data analysis will be performed using standard statistical and time-to-event methods, as well as routinely applied AI techniques. We will work with University partners (e.g. Bayes Forum) on cutting-edge AI methods and methods such as natural language processing, and similarly with HDR UK and the Turing.
Fundamental to our ongoing and future ambitions to provide detailed bulk and spatial multi-omic and digital pathology characterisation of our patient samples and preclinical models is development of data integration and analysis platforms. This will enable us to understand tumour biology in sufficient depth to interrogate and understand the biology of disease progression, therapeutic response and enable target identification and validation. These approaches are also critical for disease positioning our pre-clinical models (e.g. Jackstadt et al. Cancer Cell 2019) to facilitate clinically relevant preclinical trials prior to clinical implementation. The Centre will strengthen support for data scientists to analyse molecular pathology data gathered from our key Programmatic Themes initially focusing on CRC and HPB.